Combining data to build large trees
To create a framework to integrate fossil and extant data on mammalian dietary strategies I will be building and time-calibrating phylogenies of the clades of interest through the combination of molecular and morphological data. I am currently excited by the potential of the new methods available in MrBayes 3.2.1.
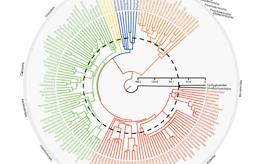
Time-calibrated trees of extinct & extant mammals
Part of the supermatrix used to build the perissodactyl (odd-toed hoofed mammals) tree.

I focus on techniques for building and time-calibrating large trees from previously published data by combining either character data such as DNA sequences (supermatrices) or tree topologies (supertrees). These trees facilitate my research into large-scale trends in vertebrate radiations as I need robust phylogenies of complete clades to act as a framework for comparative analyses.
The pro’s and con’s of supertrees that combine tree topologies, compared to supermatrices that combine raw character data (genetic and also potentially morphological) are hotly debated.Recently, myself and my colleague Olaf Bininda-Emonds used a small clade of mammals known as the Perissodactyla (odd-toed hoofed mammals) to compare the supertree (combining tree topologies) and supermatrix (concatenating character data) tree building methods using a molecular dataset. Using all the available gene sequence data on GenBank we created a multi-gene dataset. We analysed this dataset in two ways, firstly by concatenating it to make a supermatrix then using Bayesian methods to estimate the tree topology and secondly by building individual trees for each gene and combining the topologies using matrix representation with parsimony (MRP).We found that in general both the supertree and supermatrix approaches performed equally well when analysing an identical dataset; both were hindered by the current paucity of molecular data and its patchy distribution within the horse/zebra/donkey clade.
Price, S.A. & Bininda-Emonds, O.R.P. 2009 A comprehensive
phylogeny of extant horses, rhinos and tapirs (Perissodactyla)
through data combination. Zoosystematics and Evolution 85(2),
277-292.
Comparing supertrees & supermatrices
Previously I worked with a group of collaborators to build a
complete tree of extant mammals (see here) and during the process
we also investigated various methodological issues when building
supertrees, including devising a protocol to minimise the use of
poor quality and redundant data that might adversely impact the
topologies generated by supertree methods .
Bininda-Emonds, O. R. P., Jones, K. E., Price, S. A., Cardillo, M., Grenyer, R. & Purvis, A. Garbage in, garbage out: data issues in supertree construction. 2004. In Phylogenetic Supertrees: Combining Information to Reveal the Tree of Life. (ed. O. R. P. Bininda-Emonds), pp. 267-280. Kluwer Academic, Dordrecht.
Bininda-Emonds, O. R. P., Jones, K. E., Price, S. A., Grenyer, R.,
Cardillo,M., Habib, M., Purvis, A. & Gittleman, J. L. 2003.
Supertrees are a necessary not so-evil: A comment on Gatesy et al.
Systematic Biology 52, 724-729.
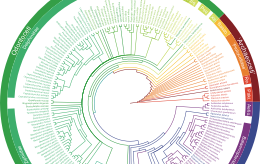
Supertree methods & best practices
Supertree of extant mammals, generated from the collaborative project. Figure 1 from Bininda-Emonds et al. 2007.
X Contact: saprice at ucdavis dot edu, Department of Evolution & Ecology, 1 Shields Avenue, UC Davis, CA 95616
X Contact: saprice at ucdavis dot edu, Department of Evolution & Ecology, 1 Shields Avenue, UC Davis, CA 95616